8. kernex#
import jax
import jax.numpy as jnp
import kernex as kex
import matplotlib.pyplot as plt
@kex.kmap(kernel_size=(3,))
def sum_all(x):
return jnp.sum(x)
x = jnp.array([1, 2, 3, 4, 5])
print(sum_all(x))
[ 6 9 12]
@kex.kscan(kernel_size=(3,))
def sum_all(x):
return jnp.sum(x)
x = jnp.array([1, 2, 3, 4, 5])
print(sum_all(x))
[ 6 13 22]
@jax.jit
@kex.kmap(kernel_size=(3, 3, 3), padding=("valid", "same", "same"))
def kernex_conv2d(x, w):
# JAX channel first conv2d with 3x3x3 kernel_size
return jnp.sum(x * w)
@kex.kmap(
kernel_size=(3, 3), padding="valid", relative=True
) # `relative`= True enables relative indexing
def laplacian(x):
return (
0 * x[1, -1]
+ 1 * x[1, 0]
+ 0 * x[1, 1]
+ 1 * x[0, -1]
+ -4 * x[0, 0]
+ 1 * x[0, 1]
+ 0 * x[-1, -1]
+ 1 * x[-1, 0]
+ 0 * x[-1, 1]
)
print(laplacian(jnp.ones([10, 10])))
[[0. 0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0.]]
@kex.kmap(kernel_size=(3, 3), relative=True)
def identity(x):
return x[0, 0]
@jax.jit
@kex.kmap(kernel_size=(3, 3), padding="same")
def get_3x3_patches(x):
# returns 5x5x3x3 array
return x
mat = jnp.arange(1, 26).reshape(5, 5)
print(mat)
[[ 1 2 3 4 5]
[ 6 7 8 9 10]
[11 12 13 14 15]
[16 17 18 19 20]
[21 22 23 24 25]]
print(get_3x3_patches(mat)[0, 0])
[[0 0 0]
[0 1 2]
[0 6 7]]
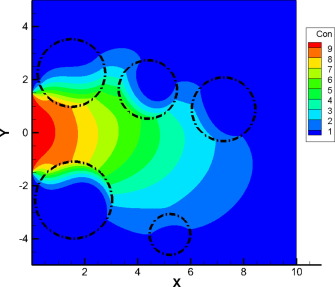
# # see https://nbviewer.org/github/barbagroup/CFDPython/blob/master/lessons/01_Step_1.ipynb
# tmax,xmax = 0.5,2.0
# nt,nx = 151,51
# dt,dx = tmax/(nt-1) , xmax/(nx-1)
# u = jnp.ones([nt,nx])
# c = 0.5
# # kscan moves sequentially in row-major order and updates in-place using lax.scan.
# F = kernex.kscan(
# kernel_size = (3,3),
# padding = ((1,1),(1,1)),
# # n for time axis , i for spatial axis (optional naming)
# named_axis={0:'n',1:'i'},
# relative=True
# )
# # boundary condtion as a function
# def bc(u):
# return 1
# # initial condtion as a function
# def ic1(u):
# return 1
# def ic2(u):
# return 2
# def linear_convection(u):
# return ( u['i','n-1'] - (c*dt/dx) * (u['i','n-1'] - u['i-1','n-1']) )
# F[:,0] = F[:,-1] = bc # assign 1 for left and right boundary for all t
# # square wave initial condition
# F[:,:int((nx-1)/4)+1] = F[:,int((nx-1)/2):] = ic1
# F[0:1, int((nx-1)/4)+1 : int((nx-1)/2)] = ic2
# # assign linear convection function for
# # interior spatial location [1:-1]
# # and start from t>0 [1:]
# F[1:,1:-1] = linear_convection
# kx_solution = F(jnp.array(u))
# plt.figure(figsize=(20,7))
# for line in kx_solution[::20]:
# plt.plot(jnp.linspace(0,xmax,nx),line)
def gaussian_blur(image, sigma, kernel_size):
x = jnp.linspace(-(kernel_size - 1) / 2.0, (kernel_size - 1) / 2.0, kernel_size)
w = jnp.exp(-0.5 * jnp.square(x) * jax.lax.rsqrt(sigma))
w = jnp.outer(w, w)
w = w / w.sum()
@kex.kmap(kernel_size=(kernel_size, kernel_size), padding="same")
def conv(x):
return jnp.sum(x * w)
return conv(image)
@jax.jit
@jax.vmap
@kex.kmap(kernel_size=(3, 3), padding=("same", "same"))
def kernex_depthwise_conv2d(x, w):
return jnp.sum(x * w)
h, w, c = 5, 5, 2
k = 3
x = jnp.arange(1, h * w * c + 1).reshape(c, h, w)
w = jnp.arange(1, k * k * c + 1).reshape(c, k, k)
print(kernex_depthwise_conv2d(x, w))
[[[ 128 202 241 280 184]
[ 276 411 456 501 318]
[ 441 636 681 726 453]
[ 606 861 906 951 588]
[ 320 436 457 478 280]]
[[1872 2770 2863 2956 1936]
[2802 4128 4254 4380 2856]
[3237 4758 4884 5010 3261]
[3672 5388 5514 5640 3666]
[2304 3364 3439 3514 2272]]]
@jax.vmap # vectorize over the channel dimension
@kex.kmap(kernel_size=(3, 3), strides=(2, 2))
def avgpool_2d(x):
# define the kernel for the Average pool operation over the spatial dimensions
return jnp.mean(x)
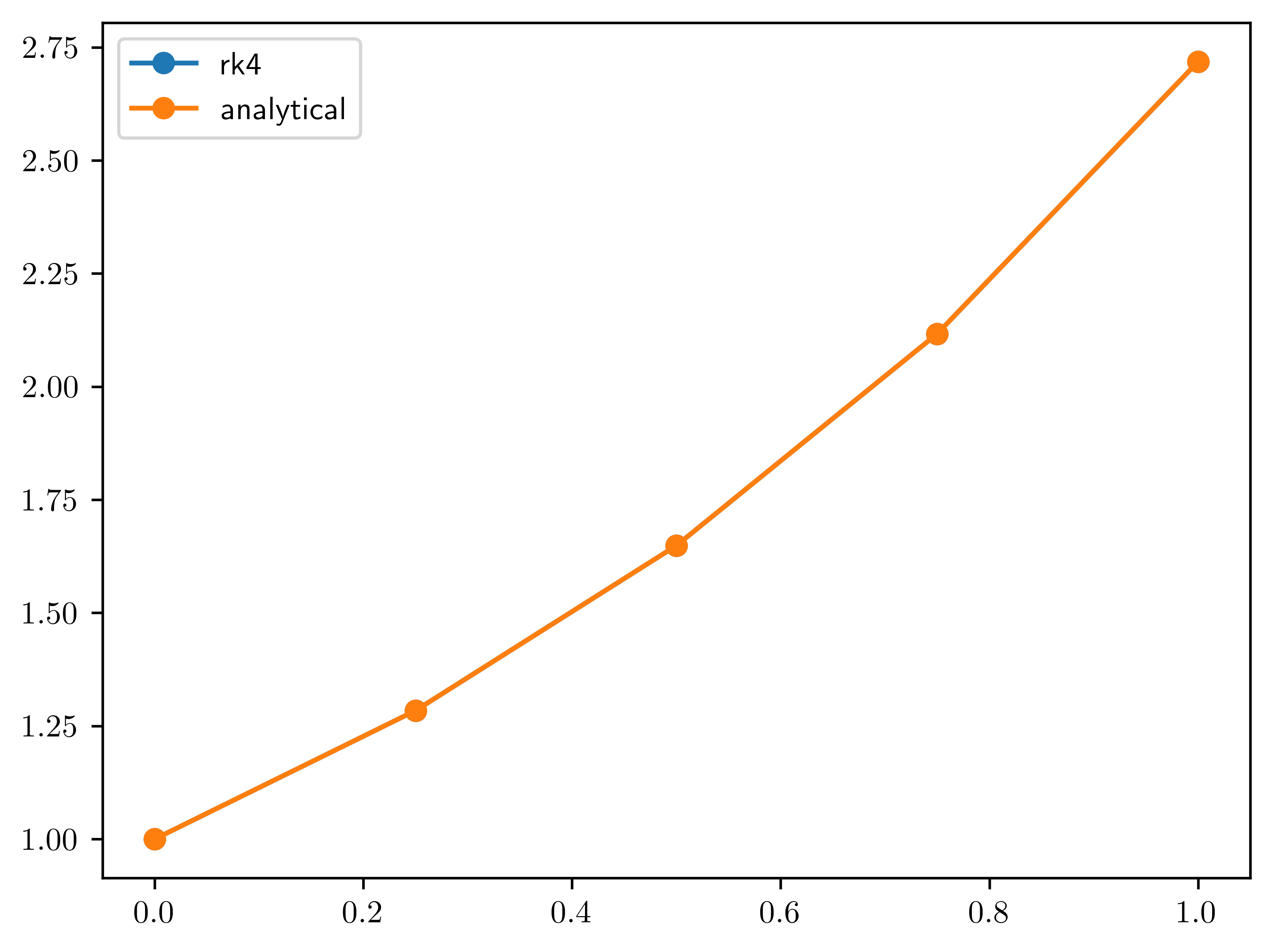
# lets solve dydt = y, where y0 = 1 and y(t)=e^t
# using Runge-Kutta 4th order method
# f(t,y) = y
t = jnp.linspace(0, 1, 5)
y = jnp.zeros(5)
x = jnp.stack([y, t], axis=0)
dt = t[1] - t[0] # 0.1
f = lambda tn, yn: yn
def ic(x):
"""initial condition y0 = 1"""
return 1.0
def rk4(x):
"""runge kutta 4th order integration step"""
# ┌────┬────┬────┐ ┌──────┬──────┬──────┐
# │ y0 │*y1*│ y2 │ │[0,-1]│[0, 0]│[0, 1]│
# ├────┼────┼────┤ ==> ├──────┼──────┼──────┤
# │ t0 │ t1 │ t2 │ │[1,-1]│[1, 0]│[1, 1]│
# └────┴────┴────┘ └──────┴──────┴──────┘
t0 = x[1, -1]
y0 = x[0, -1]
k1 = dt * f(t0, y0)
k2 = dt * f(t0 + dt / 2, y0 + 1 / 2 * k1)
k3 = dt * f(t0 + dt / 2, y0 + 1 / 2 * k2)
k4 = dt * f(t0 + dt, y0 + k3)
yn_1 = y0 + 1 / 6 * (k1 + 2 * k2 + 2 * k3 + k4)
return yn_1
F = kex.kscan(kernel_size=(2, 3), relative=True, padding=((0, 1))) # kernel size = 3
F[0:1, 1:] = rk4
F[0, 0] = ic
# compile the solver
solver = jax.jit(F.__call__)
y = solver(x)[0, :]
# %config InlineBackend.figure_format='retina'